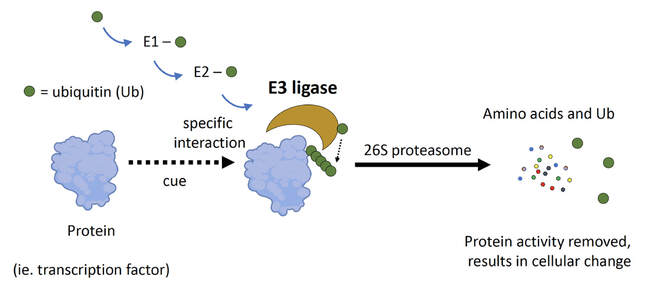
A defining, and perpetually intriguing, feature of plants is their ability to thrive in dynamic environments. External changes, in particular stressful conditions, stimulate responses in plants that help increase resiliency, prolong survival, and maximize reproductive success. Organism-level changes result from cell-level events that almost universally require selective protein degradation by the ubiquitin 26S proteasome system (UPS). Removing specific regulatory proteins from a cell triggers the activation cellular pathways associated with survival. Protein targets to be degraded by the UPS are marked by covalent attachment of ubiquitin, leading to recognition and degradation by the 26S proteasome (below). A three-part enzyme cascade accomplishes tagging where the last component, an E3 ubiquitin ligase, specifically interacts with and ubiquitylates the target protein, essentially marking it for death.
|
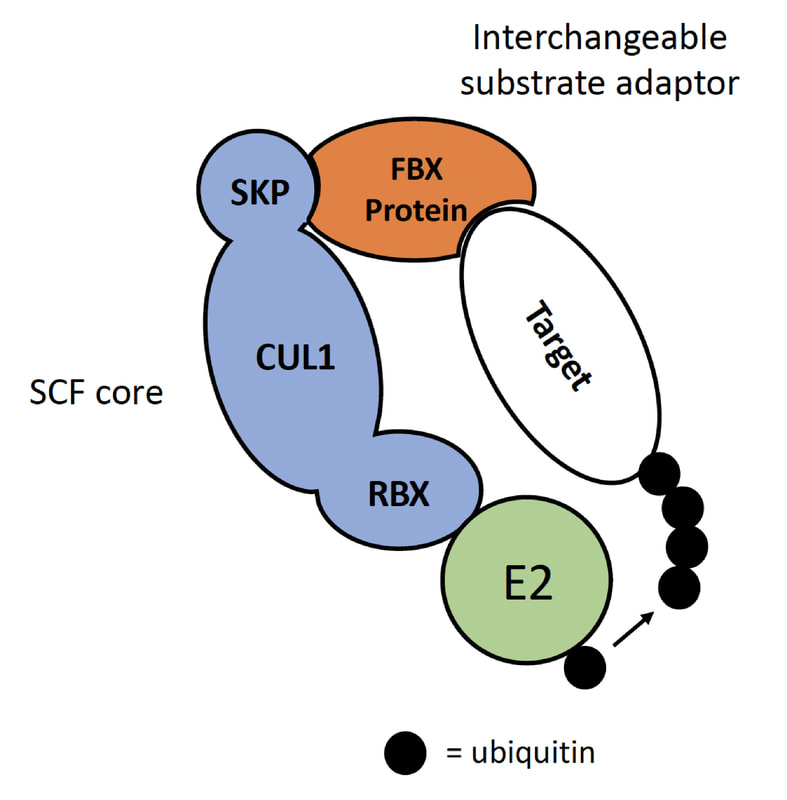
A prevalent type of E3 ligase is the SCF complex. In SCF complexes, SKP, CUL1, and an RBX protein form the common SCF core. However, F-box (FBX) proteins, which are substrate adaptors, vary between complexes and each recruit unique targets. Astoundingly, plant genomes are enriched in FBX genes by about an order of magnitude compared to most non-plant genomes. For example, there are 698, 764, and 359 FBX genes in Arabidopsis (a model plant), rice, and corn, respectively; but yeast, fruit fly, and humans contain only 20, 27, and 69, respectively. FBX proteins directly control essential plant processes, such as hormone signaling, defense against pests and pathogens, circadian rhythms, and development. Thus, FBX genes comprise a fundamentally important gene family in plant biology. However, despite FBX proteins being “center-stage” in many essential plant biological processes, about 85% of FBX genes in Arabidopsis have unknown function!
At left: An FBX protein (orange) interacts with specific target (white) and core of SCF complex (blue) to facilitate transfer of ubiquitin from E2 (green). |
Central goals of our research group are to:
1) Discover biological function within the large fraction of unstudied FBX genes
2) Understand how FBX proteins enable plant environmental stress responses
Our work is multifaceted; we use complementary approaches and methods to ask our scientific questions:
1) Discover biological function within the large fraction of unstudied FBX genes
2) Understand how FBX proteins enable plant environmental stress responses
Our work is multifaceted; we use complementary approaches and methods to ask our scientific questions:
|
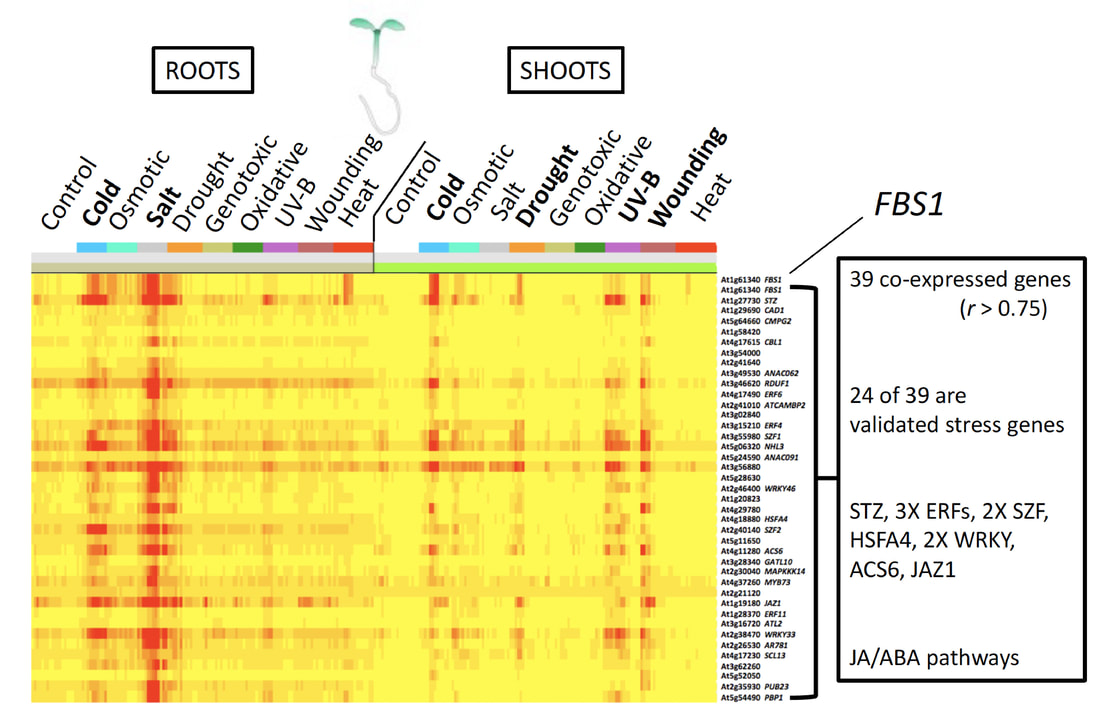
We use GENOMICS and BIOINFORMATICS approaches to identify unstudied FBX genes that respond to stress in publicly available transcriptome datasets. These candidate FBX genes are then investigated further for their roles in regulating plant responses to adverse environments.
At right: Stress-induced FBX gene FBS1 is significantly co-expressed with other known stress genes in Arabidopsis shoots and roots under abiotic stresses (red = higher expression). See Gonzalez et al., 2017. |
|
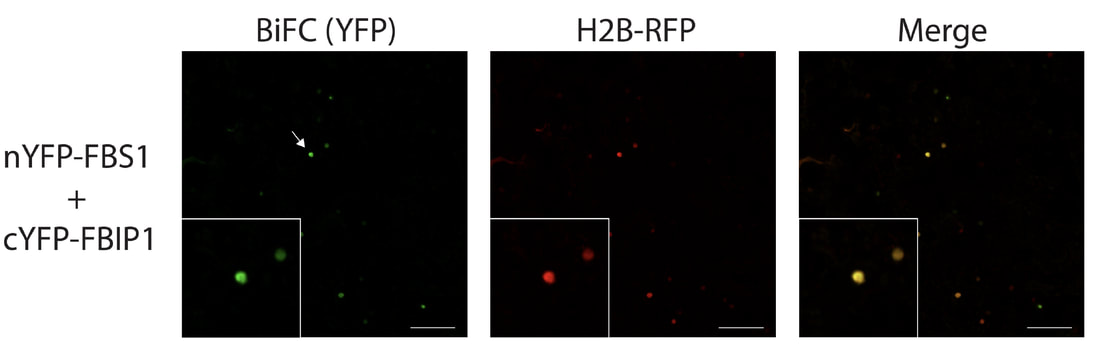
We use BIOCHEMICAL and MOLECULAR approaches to understand how FBX proteins work plant cells; we discover molecular partners for FBX proteins and identify potential ubiquitylation targets.
At left: Bimolecular fluorescence complementation (BiFC) using confocal microscopy shows that FBX protein FBS1 interacts with another protein (FBIP1) in the nucleus of plant cells. FBIP1 is a WD40 repeat-like protein of unknown function, but is highly conserved across all plant species. See Sepulveda-Garcia et al., 2021. |
|
We use GENETIC and PHYSIOLOGICAL approaches to understand how candidate FBX genes control overall plant growth and help plants survive in adverse conditions. At right: Wild type plants (left three) and plants over-expressing a candidate FBX gene (right three) hypothesized to be involved in plant growth responses under stress. |